Those studying mutations of the COVID-19 causing SARS-CoV-2 virus since its emergence in late 2019 suggest that the virus has gone through continuous evolution as it spreads into newer populations around the world.
The news of a new strain of the SARS-CoV-2 virus called D614G becoming more dominant due to the evolutionary cycle is being widely studied to understand the evolution of the virus, and what it would take for the human population to ultimately defeat it.
Read more: Immune system is able to identify COVID-19 because of common cold causing coronaviruses
As it stands, more than 22 million infections and over 788,000 deaths globally signals that the virus has anything but slowed down in the last eight months or so, showing that just as there are measures being taken to stop its spread, the virus is also mutating continually to survive and infect more and more people.
A study being performed in India has looked at the various mutations the SARS-CoV-2 virus has spawned after adapting to the geography of the area and found ways to infect people. Research conducted at the Universities of Bath and Edinburh has found over 6,000 mutations after analysing more than 15,000 genomes of the SARS-CoV-2 virus.
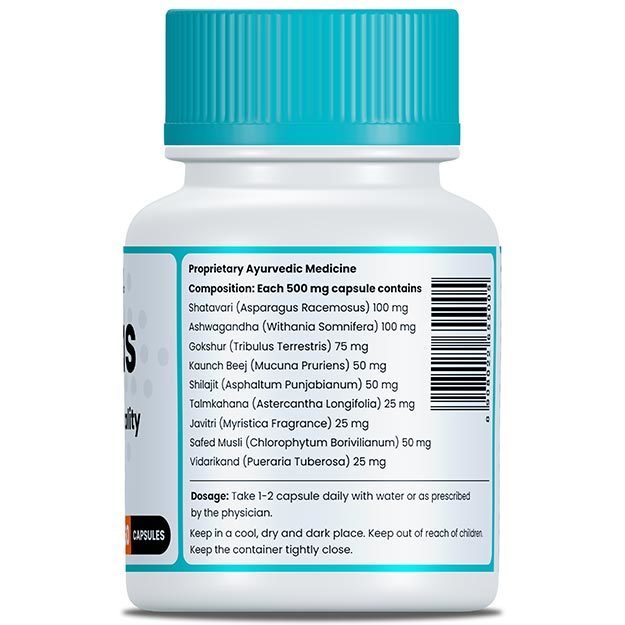
The study, performed by researchers in Kolkata and Kerala and is available on the preprint server bioRxiv, attempts to find out the evolutionary changes in the SARS-CoV-2 virus through its various mutations in India has played a role in the declining fatality rate during the period of the study (between 11 April and 28 June 2020).